[Javascript needed]
[Javascript needed]

ASSP Facts about climate changeFacts about climate changeCambiamento climaticoClimate changeParents For Future Heidelberg |
|
Seqool - A sequence analysis tool
Signal search, pattern recognition, and sequence statistics
Seqool is a free (for educational use) sequence analysis software designed primarily for searching biological signals in nucleic acid sequences. The sequence analysis program package provides several pattern recognition models, but it also includes the most common sequence analysis statistics, such as GC content, codon usage, etc.
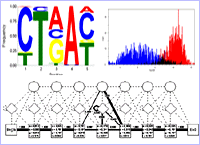
The Seqool sequence analysis software offers several pattern recognition methods for searching for biological signals, such as splice sites or user specific signals. Pattern recognition models include weight matrices (profiles), position specific score matrices, binding energy based signal search models (e.g. for snRNAs), maximum dependence decomposition models, profile hidden Markov models, and models scorring the composition of a sequence. Models can be combined using e.g. decision trees or neural networks in order to construct more refined models or for combining information from several models or sequence statistcs for a final classification of a signal.
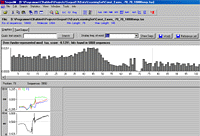
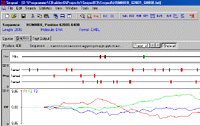
The Seqool sequence analysis tool also includes text search algorithms for the identification of simple signals. The support of IUPAC nucleic acid codes (such as ‘y’ for pyrimidines) allows less strict text searches. Over-represented oligonucleotides ("words") can be identified and optionally clustered to groups of similar words. Additional features of the sequence analysis software include the calculation of sequence composition statistics (GC, codon usage, nucleotide and oligonucleotide frequencies) and a manipulation and extraction tool for sequence or text-files.
The main features of the Seqool sequence analysis program package
Basic sequence analysis:
Signal search:
Combination of models:
File format support:
Features • Manual • Download • Contact/imprint • Privacy policy
BIOSSC/Seqool is not responsible for the content of external links.
Last Changes 24.02.2007